Processing Multimedia Biomedical Information for Disease Evolution Monitoring
by Sara Colantonio, Maria Grazia Di Bono, Gabriele Pieri and Ovidio Salvetti
A methodology for the automatic monitoring of diseases using biomedical data in multimedia is proposed. We adopt a computational intelligence approach mainly based on a multilevel neural network architecture. This approach has been employed in applications for neurosignal and image categorisation.
Models that monitor disease evolution are often based on approaches to diagnosis and prediction that compare the information obtained from a set of diagnostic exams against a set of reference parameters. A set of rules then derives a prediction of the actual and future state of health of the patient. Such models are very rigid and do not easily adapt to variations in the evolution of diseases under different and/or aleatory conditions.
There are a number of other approaches to the monitoring of disease evolution. Model-based diagnostic approaches need accurate domain models and require a fixed number of diagnostic classes, which thus reduces their flexibility. In case-based approaches, the expert's knowledge is stored in a library of cases; however, the search for the best-matching case can be computationally expensive. Inductive learning, which can include decision trees, statistical classifiers and neural networks is also used.
We are currently studying a new methodology that belongs to the field of computational intelligence. Certain diseases can be monitored automatically by processing correlated biomedical data in different media. In particular, we are developing an innovative model for the realization of a valid diagnosis methodology that can also provide an evaluation of the course of the monitored disease, based on a Multilevel Artificial Neural Network (MANN) architecture.
Multimedia information related to diagnostic and/or therapeutic processes can be of different types and comes from several sources:
- images (eg CT scan, MR scan, etc.)
- signals (eg EEG, pressure, etc.)
- historical and clinical data
- laboratory data (eg blood or specimen analyses, etc.).
We are thus designing and developing a computational method to support disease diagnosis and evolution prediction that can process these different types of biomedical data.
For any given case under examination, a set of parameters, consisting of features extracted from the multimedia data available, is processed through a MANN. The MANN is organised in three hierarchical levels corresponding to three processing phases:
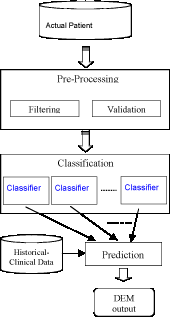
- pre-processing phase, dedicated to input data filtering and validation to eliminate incongruencies and to control consistency
- classification phase, performed on the output of the previous phase to identify the actual state of the disease
- prediction phase, to evaluate the course of the disease using the results of the previous phase together with patient's historical and clinical data.
The multilevel approach guarantees both specialisation and adaptability, since the single levels of the network can be optimised according to the specific characteristics of the problem to be solved, and the entire architecture can be easily integrated if the problem description changes, by simply training only those levels involved in the modification.
The advantages of using a MANN architecture can be summarised in two main points: the modular organisation facilitates analysis of the networks and the hierarchy enables the network to finely tune itself towards recognising the most promising directions to look for relevant patterns.
This multilevel architecture is shown in Figure 1. Multimedia data on the patient are acquired through several diagnostic modalities and passed to the pre-processing phase. This phase is dedicated to the selection of the relevant parameters (filtering) and to their validation with respect to known reference data. The pre-processed data are classified in the second phase in order to evaluate the patient's current state. The final phase provides a prediction of the evolution of the disease, using two different sets of data: the parameters obtained from the classification level, and historical and clinical data for other patients and diseases.
|
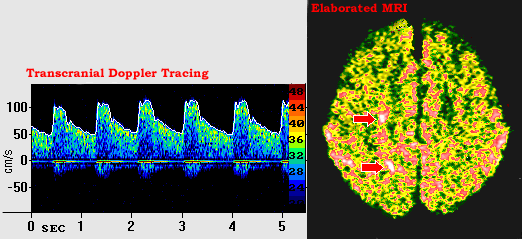
Our model has been applied to a real case study of a brain pathology, selected because of its clinical interest. Using a set of multimedia data composed of transcranial doppler (TCD) ultrasound signals, magnetic resonance (MR) neuro-images and other clinical data, the model has been instantiated for the monitoring of carotid and cardiovascular steno-occlusive diseases and the diagnosis and follow-up of cerebral anomalies. Figure 2 shows some of the information used to monitor this patient.
|
From the results of our first experiments, this model appears to be an effective tool for supporting the diagnosis activity and work is underway to integrate it into a hybrid system for medical decision support.
Please contact:
Sara Colantonio or Maria Grazia Di Bono, ISTI-CNR, Italy
Tel +39 050 315 3146
E-mail: Sara.Colantonio![]() isti.cnr.it, Maria.Grazia.Dibono
isti.cnr.it, Maria.Grazia.Dibono![]() isti.cnr.it
isti.cnr.it

 This issue in
This issue in