Towards an Integrative and Context-Sensitive Approach to In Silico Disease Modelling
by Matej Oresic, Peddinti V. Gopalacharyulu, Erno Lindfors, Catherine Bounsaythip, Ilkka Karanta, Mikko Hiirsalmi, Lauri Seitsonen and Paula Silvonen
Historically, the scientific methods applied to biological problems have largely been limited due to the fact that it has been difficult to collect the data. Today, these scientific methods are challenged by the 'omics' revolution, which are empowering us with the ability to collect large amounts of data in parallel from a particular system. However, the development of efficient tools to exploit this data within the context of biological systems has been much slower. Due to this mismatch the knowledge acquisition in life sciences is actually increasingly difficult and new information technology solutions are needed to resolve this problem.
The overall objective of the project is to build an integrated system with explanatory and predictive abilities to represent biological and clinical level knowledge related to human diseases. In order to achieve sufficient focus, we will initially limit the project to domains of type II diabetes, cardiovascular complications associated with metabolic syndrome, and obesity. The project is a collaboration between the two VTT units, Information Technology and Biotechnology. We will also cooperate with other national and international experts in specific disease domains.
The primary reason for pursuing this goal is that a vast yet dispersed amount of information already exists about these diseases, and with advancement of new high-throughput technologies the amount of information is rapidly increasing. But this information will become knowledge only when it is mapped to a certain knowledge structure, ie, organized or linked together in a way that makes it accessible or interpretable by the users. In fact, the same pieces of information can yield different knowledge depending on the context and purpose,. Moreover, context-sensitivity means also that the 'emerged' concepts should match the community's consensus, ie, people working in the same field.
Project Structure
Our project can be divided into several sub-projects:
- develop multilevel bioinformatics data management framework based on XML and Semantic Web technologies
- develop software solutions to map the information to a knowledge structure, including the use of text annotation, common vocabularies and semantics
- implement a data and text mining solution for mining quantitative and qualitative relationships between entities of interest
- select most salient concepts in respect to different levels of the biological system to model disease, and set up an a priori framework for the ontology model linking the levels
- populate the model within the ontology framework, modify the model and framework and add new entities through validation.
In longer term, we wish to pursue the following goals:
- develop methodology to create predictive quantitative and dynamical models from the knowledge base
- validate the modelling tools by applying them to predict responses to specific interventions for which sufficient data already exists.
Handling Structured Data
Much of biological information, in particular at biomolecular level, is stored in databases, such as genome databases (eg GenBank), pathway databases (eg KEGG), protein databases (eg UniProt) or small ontologies like Gene Ontology (GO). Even at this level, a continuous frustration of researchers is the fact that it is not easy to map identical entities across multiple databases, due to different naming conventions. Therefore, any attempt at data integration should start with identifying the 'atoms of information' and creating solutions to resolve the names. XML and RDF are useful technologies for creating such identity-mappings across multiple data sources. There are ongoing efforts to standardise the life science data formats in order to facilitate the exchange of information and knowledge (eg W3C Semantic Web Life Science). For every database (either containing annotations or information about entity relationships) we create a simple XML schema that enables mapping to other databases.
Textual Data and Concepts
Most of life science information is still available only in textual form. This is particularly true for information on relationships between the molecular level events and more complex concepts such as events related to a multifactorial disease. Text mining solutions are therefore needed to sift through the semi-structured disease-related data such as OMIM, literature (eg PubMed) and patent databases. Established vocabularies and concepts such as UMLS may be of help, but one should be aware that with rapid progression of life sciences and medicine today the new terms and concepts are rapidly emerging. There have already been efforts in the text mining area to infer knowledge from available sources in the domain, for instance the BioCreAtIvE community is set up to assess current text mining tools in the area of biology.
In our project, we use GATE as software platform, augmented with various scripts and programs. We extract (subject, predicate, object) triplets from the raw text mass. We use a seed vocabulary of terms – at least two terms of the triplet must be found in the vocabulary – and the vocabulary is then augmented with the third term, if needed. The extraction is based on shallow parsing: instead of forming complete parse trees from each sentence, we rather extract noun phrase and verb phrase blocks that are further processed to produce the triplets. In the end, the combination of all these triplets will form a conceptual graph that tells about the relations between the entities of interest.
Everything is Connected!
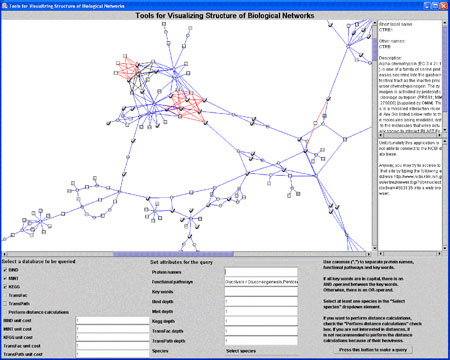
As a start towards these goals, VTT BEL has already integrated data from UniProt, KEGG, TransFac, TransPath, MINT, and BIND databases using XML; and developed a Java-based tool that allows parallel retrieval across multiple databases, including metabolic pathways, protein-protein interactions, signaling and regulatory networks. The results are then visually displayed as a network (see figure). Edge attributes contain information about type of relationship, possibly quantitative or semantic information (such as 'is located in' in case of linking a protein with a complex entity such as cell organelle, with information obtained by text mining).
|
Our aim is to connect entities (nodes), which can include molecular entities as well as more complex concepts such as insulin resistance or diabetes, with relationships (edges) which can either be direct physical interactions or more complex relationships. Nodes and edges may be clustered and mapped to an ontology-type structure. Specifically, we are also interested in retrieving quantitative information on relationships which can be used for predictive modeling in the future.
Multilevel Inference
Finding linkages between different entities in multifactorial diseases is a demanding task due to multiple levels of biological organization being involved in disease pathogenesis and progression. At the first phase of the project we want to accumulate and integrate the data on entities and relationships among them, which can be used for explanatory modelling, ie, for interpretation of results from experiments using clinical or genomic screening, transcriptional, protein, or metabolite profiling. Once we obtain the networks of associated entities across different levels, the inferences will be made based on the network's topology. Therefore, our initial computational efforts will focus on studies of network topologies and their associations with known experimental data and physiological conditions.
Links:
VTT: http://www.vtt.fi/
Quantitative Biology and Bioinformatics group at VTT BEL: http://sysbio.vtt.fi/qbix/
GenBank: http://www.ncbi.nlm.nih.gov/
UniProt: http://www.ebi.uniprot.org/index.shtml
Kyoto Encyclopedia of Genes and Genomes: http://www.genome.jp/kegg/
Gene Ontology: http://www.geneontology.org/
W3C Semantic Web for Life Sciences: http://lists.w3.org/Archives/Public/public-semweb-lifesci/
PubMed: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi
Online Mendelian Inheritance in Man: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM
Unified Medical Language System: http://www.nlm.nih.gov/research/umls/
BioCreAtIvE: http://www.pdg.cnb.uam.es/BioLINK/BioCreative.eval.html
GATE: http://gate.ac.uk/
Please contact:
Matej Oresic, VTT Biotechnology, Finland
Tel: +358 9 456 4491
E-mail: matej.oresic![]() vtt.fi
vtt.fi

 This issue in
This issue in