Biomedical Imaging for Enhanced Genetic Data Analysis
by Thanasis Margaritis, Kostas Marias, Manolis Tsiknakis and Dimitris Kafetzopoulos
The Institute of Molecular Biology and Biotechnology in collaboration with the Institute of Computer Science - FORTH, have initiated several joint efforts towards the development and implementation of advanced techniques for biological data analysis and management. One of the most promising research areas is biomedical imaging, a multidisciplinary field aiming at effective analysis and processing of biological/genetic data. It can also be considered as one of the emerging areas in biomedical informatics, emanating from synergies amongst medical informatics, medical imaging and biology. The aim of this field is to build on previous engineering knowledge, in order to develop robust analysis frameworks for maximizing the information content of biological data.
Traditionally, medical imaging has focused in providing anatomical information, mainly imaging human bones, dense tissue and arteries. Several advances, especially positron emission tomography (PET) and functional magnetic resonance imaging (MRI), allowed the study of various pathophysiological processes via radiolabeled tracers (PET) or pharmacokinetic models in contrast enhanced MRI. Current gamma camera, PET, MRI, and optical imaging paradigms have been demonstrated for monitoring molecular-genetic processes (via direct and indirect approaches), rather than anatomy. At the same time, newly developed imaging techniques provide very important information regarding gene and protein expression and function. Some of the most interesting techniques used include:
- Microarray imaging: An array of DNA reporters is hybridized with labeled probes to study patterns of gene expression. It is based on confocal laser microscopy.
- Fluorescence in situ hybridization (FISH) techniques: A method for determining the cytogenetic location of a cloned fluorescent-labeled segment of DNA. The site of chromosomal DNA-probe hybridization is determined by fluorescent microscopy.
- Spectral karyotyping (SKY) or multiplex fluorescence in situ hybridization (M-FISH) of human chromosomes.
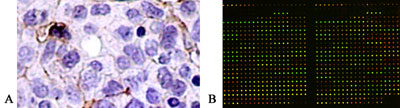
- ImmunoHistoChemistry (IHC): A method of detecting the presence of specific proteins in cells or tissues.
Here we describe several common analysis issues of microarray, FISH and IHC image-data analysis. In particular, microarray analysis is one of the first domains that clearly addressed the need to develop synergies that extended beyond the limits of previous fields, such as medical imaging and bioinformatics. Thus, gene expression imaging can be regarded as a biomedical imaging discipline that deals mainly with the following data analysis problems:
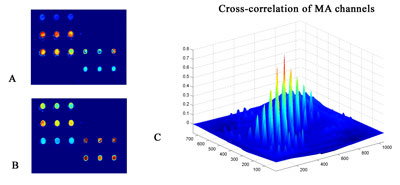
Registration: This is a typical problem for microarray imaging, as well as in M-FISH and IHC imaging, because images are collected in more than one channel. Due to the differences in the optical paths of color channels in acquisition of images and the inherent 'chromatic aberration' of the optical system, the same object in the specimen changes its off-axis position when imaged with different wavelengths. In order to align multi-spectral images a plethora of 'classic' algorithms is available. We have used an image similarity approach based on similarity measures (eg cross-correlation) in order to geometrically align microarray images(see Figure 2).
|
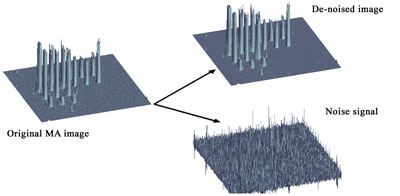
Segmentation: It is particularly crucial to be able to define the regions of interest both in microarray imaging and in FISH/M-FISH/IHC. In FISH imaging it is crucial to segment dot signals that are used then to quantify the gene duplication or deletions caused by chromosome abnormalities. Similarly, it is important to segment microarray spots from the background in order to define the relative statistical distributions of values and perform normalization. Popular methods derived from pattern recognition/neural networks include k-mean, fuzzy clustering and Markov random fields. We have used wavelet filtering as segmentation pre-processing step by separating the signal from noise (see Figure 3).
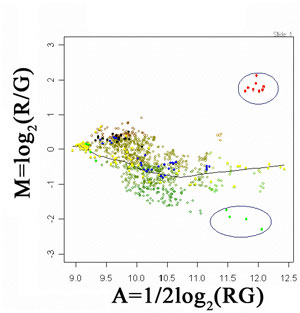
Image Feature Quantification: Following image preprocessing steps, it is crucial to derive quantitative information related to gene expression/function. In this context, important features or morphology in images need to be extracted and quantified. In an interphase FISH-labeled cell, a number of features are measured to verify if FISH dot signals are well separated dots, overlapping dots, contiguous or spread signals, or split signal dots. This quantitative information can be related to the characterizations of gene duplication or deletions, associated with several genetic diseases (eg neuropathy). On the other hand, in microarray image analysis it is important to define the actual quantitative relationship of two information channels (eg healthy tissue RNA labeled with Cy3 and a diseased tissue RNA labeled with Cy5). This way, differential expression of genes can be detected on the basis of 'differing' from the overall trend that characterizes the relationship of gene expressions. This is illustrated in Figure 4, where a LOWESS fit has been used to estimate the overall trend of intensity pairs in a microarray experiment. This way subtle differences that can be potentially attributed to differential gene-expression are preserved (see encircled regions).
|
We presented some basic research directions in biomedical imaging, with special focus in our work on imaging of gene-expression. By developing synergies within FORTH, we aim to expand the present capabilities of biological data analysis by applying modern imaging and computational algorithms, inspired from a deep understanding of the underlying biological phenomena.
Please contact:
Dimitris Kafetzopoulos and Manolis Tsiknakis, FORTH, Greece
Tel: +30 2810 391594
E-mail: kafetzo![]() imbb.forth.gr, tsiknaki
imbb.forth.gr, tsiknaki![]() ics.forth.gr
ics.forth.gr

 This issue in
This issue in