GenoLink: Discovering Drug Target Proteins by Exploring Networks of Heterogeneous Biological Data
by Patrick Durand, Laurent Labarre, Alain Meil and Jérôme Wojcik
As ever-larger amounts of biological data are made available by genomic and post-genomic technology, the conversion of these data into valuable knowledge becomes crucial to the discovery of new drug target proteins. For this reason, the GenoStar Consortium has developed GenoLink, a software platform designed for the integration and exploration of complex biological datasets.
Genomic and post-genomic technology is producing a huge amount of heterogeneous data available for investigating the functions of proteins. Biologists are now faced with the problem of exploiting these raw data and extracting useful knowledge from them. An efficient way to perform this task consists in exploring the relationships between various kinds of biological data. As a very simple example, one can assign a function to a protein from a given organism knowing that this protein has a sequence similar to a well-known protein from another organism. In such a case, a network of objects (proteins and organisms) can be explored using relationships (eg a protein 'belongs' to an organism and a protein 'is similar' to another one). More generally, the genomic world can be viewed and explored using a complex network of biological objects and their relationships. Following this idea the GenoStar Consortium has developed GenoLink, a software platform dedicated to the exploration of complex networks of biological data. GenoLink is built on three modules that handle data modelling, data integration and data querying/visualization.
Data Modelling
GenoLink uses GenoCore to model and store networks of data. GenoCore is an advanced object-oriented knowledge management system developed at INRIA. Among other features, GenoCore provides two complementary entities of representation: classes and associations. GenoLink uses these to formally describe the objects and relationships within a network. As in any object-oriented system, a class or an association represents a set of objects or relationships respectively. Classes and associations can be organized in hierarchies, allowing inheritance and specialization mechanisms.
To discover new proteins of pharmaceutical interest we have set up a knowledge base describing a network of data centred on protein-protein interactions. For that purpose our data model can handle information coming from complementary sources of data, such as fully annotated genomes, pre-computed clusters of orthologous genes, functional classifications, protein domains and protein-protein interactions. It is worth noting that this data model can be updated to accommodate new sources of data.
Data Integration
To supply the knowledge base with data, GenoLink comes with a set of import tasks. These are organized to perform integration within a single data space. Data integration is carried out using unique object identifiers as they appear in the source databases. Going along with the idea of an extensible data model, new import tasks can be added to GenoLink using the application-programming interface provided with the software.
Data Querying and Visualization
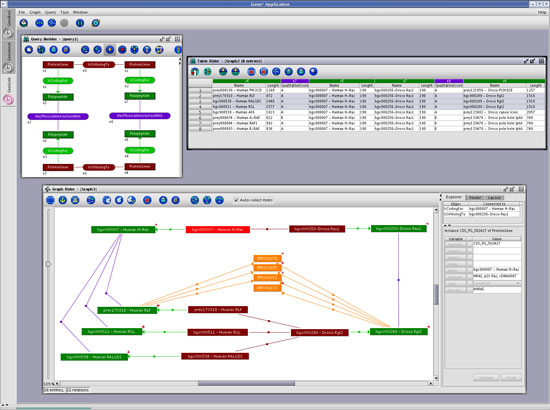
To support the exploration of complex networks of data, GenoLink has a dedicated and original query system. Queries are network patterns built up from the classes/associations defined in the data model. For a given query, GenoLink will search for matching patterns by looking for sub-networks within the full knowledge-base network. It is then possible to further explore the network starting from a query result. Through this exploration process, the biologist may be able to infer new links between previously unrelated entities.
GenoLink was entirely written using Java technology. The creation of queries and the exploration of results is achieved in an easy-to-use graphical environment. It is worth noting that GenoLink also provides a full-featured graph query language that allows the design of complex querying strategies.
Applications
In addition to the identification of proteins of pharmaceutical interest in the fields of cancer, GenoLink has been used to annotate proteins from various bacteria genomes, and to infer protein-protein interactions in several eukaryotic genomes.
|
The GenoStar Consortium
GenoLink is one of the application modules of GenoStar, a bioinformatics platform for exploratory genomics. Other application modules are GenoAnnot (for the annotation of bacterial genomes) and GenoBool (a data mining application). The development of the platform was initiated by the GenoStar consortium at the end of 1999. Partly supported by the French Ministry of Research, it brings together four partners: two biotech companies - Hybrigenics (Paris) and Genome Express (Grenoble) - and two research institutes - the Pasteur Institute (Paris) and the INRIA Rhône-Alpes (Grenoble). The GenoStar platform is now maintained by the private company GenoStar. The platform remains freely available for the academic community.
Link:
http://www.genostar.org
Please contact:
Patrick Durand, IRISA-INRIA,France
Tel: +33 2 9984 7321
E-mail: Patrick.Durand![]() inria.fr
inria.fr
Jérôme Wojcik, Hybrigenics S.A., France
Tel: +33 1 5810 3862
E-mail: jwojcik![]() hybrigenics.com
hybrigenics.com

 This issue in
This issue in