Validation of Clustering Techniques for Microarray Gene Eexpression Data
by Nadia Bolshakova and Pádraig Cunningham
Recent advances in microarray technology have enabled the measurement of the simultaneous expression of thousands of genes under multiple experimental conditions. The methods implemented in this research may contribute to the validation of clustering results and the estimation of the number of clusters. For instance, these tools may be used for the identification of new tumour classes using gene expression profiles. The results show that this estimation approach may represent an effective tool tosupport biomedical knowledge discovery and healthcare applications.
One of our major tasks is to advance data analysis and integration capabilities in genomic expression pattern discovery and classification. It has consisted of the implementation of algorithms and tools to organise and categorise genome expression data. It has integrated and improved a number of machine learning techniques, which may aid in the identification of relevant features for diagnostic, prognostic and system biology studies. These tools may also be applied to other information management domains such as biomedical informatics. Moreover, automated discovery solutions may assist the design of novel techniques for intelligent information retrieval and knowledge and meta-knowledge representation, which are crucial aspects for the integration of information over the global network.
An important step in the analysis of gene expression data is the detection of samples or genes with similar expression patterns. The accurate classification of tumours is essential for a successful diagnosis and treatment of cancer. One of the problems associated with cancer tumour classification is the identification of unknown classes using gene expression profiles. Several clustering algorithms have been developed for gene expression data. Also techniques to systematically evaluate the quality of the clusters have been presented. The prediction of the correct number of clusters in a data set is a critical problem in unsupervised classification. Various cluster validity indices have been proposed to measure the quality of clustering results. It is useful not to rely on one single clustering or validation method, but to apply a variety of approaches. Therefore, a combination of these methods may be successfully used for the estimation of the number of clusters. It has been shown that these methods may support the prediction of the optimal partition and computational diagnosis.
|
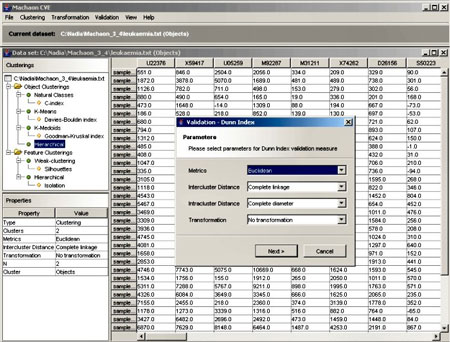
We have developed the Machaon Cluster Validation Environment (Machaon CVE) for the application of different clustering and validation algorithms to experiment on gene expression data. This tool may improve the quality of the data analysis results, and may support the prediction of the number of relevant clusters in the microarray datasets. The major stages of the system can be summarised as follows:
- Clustering. In this step we extract clusters that correspond to the pre-defined number of clusters for a particular dataset. It offers a number of the well-established clustering methods that are available in the literature as well as some recently developed ensemble techniques.
- Validation of clustering techniques. The clustering methods can find a partition in a dataset, based on certain assumptions. Thus, an algorithm may result in different clustering schemes for a dataset assuming different parameter values. Machaon evaluates the results of clustering algorithms based on quality indices and selects the clustering scheme that best fits the data. The definition of these indices is based on two fundamental criteria of clustering quality: cluster compactness and isolation.
To support biomedical knowledge representation in gene expression data analysis, a new markup language for microarray data clustering and cluster validity assessment has been developed. cluML, a free, open, XML-based format has been designed to address some of the limitations observed in traditional formats, such as inability to store multiple clustering (including biclustering) and validation results within a dataset.
To enhance the predictive reliability and biological relevance of the validation results, a knowledge-driven cluster validity assessment approach for microarray data clustering has been implemented. It consists of validity indices that incorporate similarity knowledge originating from the Gene Ontology (GO), which is a structured, shared vocabulary that allows the annotation of gene products across different model organisms.
The methods performed in this research may bring contribute to the evaluation of clustering outcome and the prediction of optimal cluster partitions. The described estimation approach represents an effective tool to support biomedical knowledge discovery in gene expression data analysis. Despite the fact that Machaon CVE was developed for DNA microarray expression analysis applications, it may be effectively used for clustering and validating of other biomedical and physical data with no limitations.
Please contact:
Nadia Bolshakova, Trinity College Dublin/IUC, Ireland
Tel: +353 1 608 3688
E-mail: Nadia.Bolshakova![]() cs.tcd.ie
cs.tcd.ie

 This issue in
This issue in