| |||||||
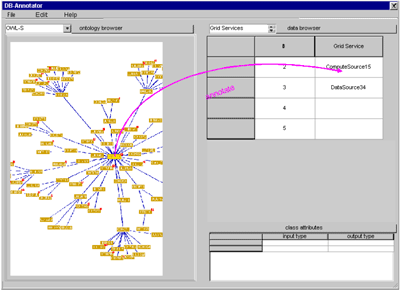
Bioinformatics in the Semantic Gridby Kai Kumpf and Martin Hofmann One of the major challenges in Grid computing is the semantics-driven retrieval of Grid services and distributed data. DB-Annotator is an annotation tool for data in Grid services developed in a collaborative project between the bioinformatics department and the department for Web applications at the Fraunhofer Institute for Scientific Computing (SCAI). Fully annotated data in the Grid is particularly important in bioinformatics. DB-Annotator was conceived for the Resource Description Framework (RDF) annotation of structured information sources such as relational data or XML-based service descriptions. Only semantically annotated Grid services (GS) provide a means of finding data or compute-services that are suitable for the task at hand. Furthermore, they make the building of workflows (coupled services within the grid) a realistic goal. DB-Annotator will support several levels of semantic annotation to enrich Grid services, ranging from the services themselves to the data within the Grid. The project DB-Annotator (see Figure 2) was designed for universal annotation of data that can be represented in tabular form, ie data that can be qualified by unique keys.
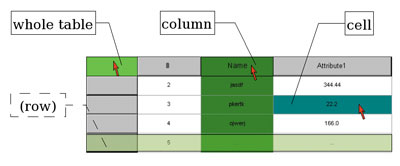
The main thrust for the development came from the realization that most interesting data within biological databases resides within free-form text fields and is therefore not easily accesible for data mining. Relational databases (RDB) are usually the only choice for data retrieval within distributed organizations. Categorial description of the typed data residing in RDBs can be derived from the tables (entities) themselves, and from the columns (entity attributes) and the cell contents (instances of attributes). Still more fine-grained information comes from the notorious ‘description’ fields, but putting this implicit information to use for machine-reasoning is unrealistic at this point. The goal was thus to provide easy navigation through existing ontologies and data-source-independent RDF annotation of RDB data via drag and drop (see Figure 2). This data categorization can take place on three levels within a database (see Figure 3), not counting the database itself. A unique data key that can be linked to an ontological category will therefore appear as ‘database:schema:table[:column:row]’, the latter two key parts being optional depending on the depth of the annotation.
This concept of assigning ontology classes to data was inspired by existing annotation software for the Semantic Web. Once a data source has received ample annotation, data mining will be possible by employing standard methods on the categorized data. With annotated RDB, access to data services in the Grid will improve significantly, since querying by categories is enabled across several RDBs at once. Further, every result set will be enhanced with the respective category information, a data basis that could be used to support further automated annotation of newly inserted data.
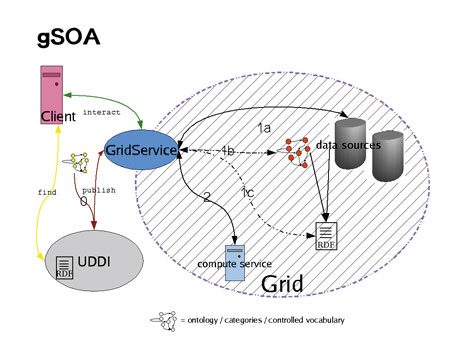
Since DB-Annotator produces standard RDF (subject-predicate-object triples like ‘entity_xyz is-a category_abc’), its output can also serve to enhance UDDI, a proposal made for the introduction of content-searchable Web services and a possible starting point for GS-workflow construction. Instead of the database unique keys, the RDF would contain qualifiers for the available Grid services. Eventually a semantically enhanced Grid Service-Oriented Architecture (gSOA – SOA being a term borrowed from the Web services world) would look like Figure 1. Full categorization of both GS and the exchanged data leaves no more room for ambiguities – a precondition for data exchange and manipulation within the Grid network of virtual organizations (VO). Given the bulk of biochemical entities that go under various names and interact on different levels, the relevance of fully annotated data Grid services in bioinformatics, especially systems biology, is enormous.. Until now, ambiguities in synonyms and interactions had to be detected by human intelligence,– meaning full-fledged systems biology services won't be successfully automated without proper categorization. Since DB-Annotator draws on a common set of agreed ontologies (eg gene ontology) for the annotation of both data services and data sets or subsets, it can provide the semantic glue on all levels required. The project was launched in the department for bioinformatics at Fraunhofer SCAI. It is a collaborative project between the bioinformatics department and the department for Web applications. Preliminary work commenced late in 2003 with the aim of producing a prototype that would capture the basic functionality of the future system. The prototype, featuring 1:n mapping of a database to several ontologies and RDF flatfile storage, was completed along with a detailed requirement analysis this summer. A new structured development from scratch has now started and is expected to be finished early in 2005. Among the things that will be included in the new phase are n:m relationships between data and ontology classes and storage in a single, structured relational repository. The product is currently intended to provide support for semantic annotation of biological and chemical databases. The possibilities for Grid services annotation within UDDI are explored within the semantic grid research group under the auspices of the Global Grid Forum (GGF). Links: Please contact: | |||||||