|
|||||
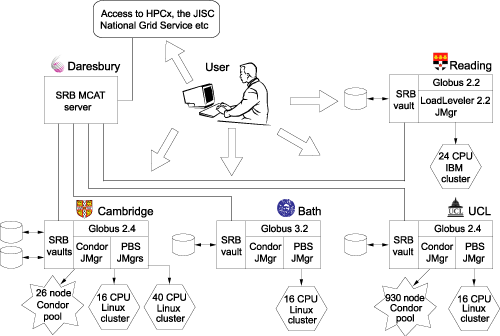
The eMinerals Minigrid: An Infrastructure to Support Molecular Simulation Scientistsby Martin Dove The eMinerals project is one of the UK Natural Environment Research Council (NERC) e-science testbed projects (formal name 'Environment from the Molecular Level'). The scientific aim is to use Grid technologies to push forward the capabilities of molecular-scale simulations for the study of environmental processes. Examples include issues such as nuclear waste encapsulation, adsorption of pollutants on the surfaces of soil particles, and the interaction of mineral surfaces and fluids. The simulations we perform range in their complexity. In some cases we need to simulate systems containing millions of atoms, and for this we represent the forces between atoms by simply parameterised models. In other cases we need to have more accurate representations of these forces, and we use quantum mechanical methods, but with smaller numbers of atoms. At its heart, the challenges faced in scaling up these calculations are computational, but with larger simulations comes an increased need manage data in new ways. In addition to the fact that larger simulations generate larger data files, we also have the problem that with higher throughput of calculations comes the need for intelligent file management within an individual study. The eMinerals minigrid has been developed as an integrated compute and data grid with the aim to equip the scientists to work with more complex simulation studies than before. The eMinerals minigrid is built around a heterogeneous set of computing resources, quite deliberately so because different types of molecular simulations have very different requirements. The compute resources include 4 clusters of PCs, an IBM p-series parallel computer, a Condor pool containing a range of machines but primarily designed to allow test jobs with high memory requirements, and a large (930 machine) Condor pool containing the teaching PCs of University College London. The latter is the largest Condor installation in the UK, and is designed to allow high-throughput calculations that do not have high memory requirements. Access to these machines is through Globus, with each member of the team having a UK digital certificate to ensure authorisation and authentication. Moreover, we expect the scientists to submit jobs using the Globus toolkit commands rather than via direct logins to the resources. The only logins are permitted to allow code developers to compile and test.
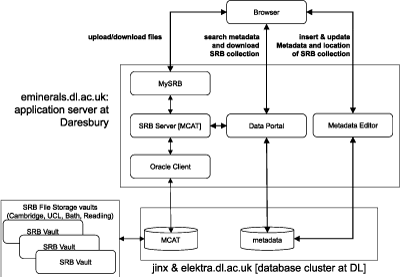
One immediate problem with this regime is that of file transfer. Both Globus and Condor present problems for users in this regard. Our solution has been to directly incorporate the Storage Resource Broker (SRB) developed by the San Diego Supercomputer Centre. The SRB provides a means to distribute files across a number of file systems (called 'SRB vaults') but to be seen by the user as a single file system. The physical location of files is transparent to the user, and is seen only as a file attribute. We have set up 5 SRB vaults across the eMinerals minigrid. The integrated compute and data grid that constitutes the eMinerals minigrid is shown in Figure 1, with the data component shown in more detail in Figure 2.
The integration of the SRB within the eMinerals minigrid facilitates the following workflow. When a user wants to run a job, the first task is to upload the relevant files to the SRB. The user then executes a three-stage job using Condor-G, which provides a Condor-style wrapping for the Globus commands. The first stage is for these files to be downloaded from the SRB onto the compute resources being used. The second stage is to run the job. The third stage is to put all output files into the same directory within the SRB for easy access to the user. These three stages have been wrapped within a single script written by one of the project members, although we are working on a compute portal to make job management easier for the scientists. The final stage in the process is longer-term management of the data. The Daresbury project partners have developed a data portal that has a direct interface with the SRB. Data that require long-term storage are uploaded to the data portal from the SRB, and provided with metadata that enables use to be made of the data by the scientist or collaborators at a later date. In order to ensure interoperability of data, we are making use of the Chemical Markup Language, one of the established XML standard languages. This report represents work in progress. Components of this work, particularly the eMinerals minigrid as an integrated compute and data grid, are now beyond proof-of-concept and are in production mode. Immediate tasks are to provide the tools such as the compute portal to give the scientists more help in their use of the full minigrid. The eMinerals team consists of scientists, applications code developers and computer scientists/grid specialists from the Universities of Cambridge, Bath, and Reading, University College London and Birkbeck College, and the CCLRC Daresbury Laboratory. Link: Please contact: |
|||||