
by Claudine Médigue, Yves Vandenbrouke, François Rechenmann and Alain Viari
‘Imagene’ is a a co-operative computer environment for the annotation and analysis of genomic sequences developed in collaboration between INRIA, Université Paris 6, Institut Pasteur and the ILOG company. The first version of this software was dedicated to bacterial chromosomes. Its capabilities are currently extended to handle both prokaryotic and eukaryotic data and to link pure genomic data to ‘post-genomic’ data, particularly metabolic and gene expression data.
In the context of large-scale genomic sequencing projects the need is growing for integration of specific sequence analysis tools within data management systems. With this aim in view, we have developed the Imagene co-operative computer environment dedicated to automatic sequence annotation and analysis (http://abraxa.snv.jussieu.fr/imagene). In this system, biological knowledge produced in the course of a genome sequencing project (putative genes, regulatory signals, etc) together with the methodological knowledge, represented by an extensible set of sequence analysis methods, are uniformly represented in an object oriented model.
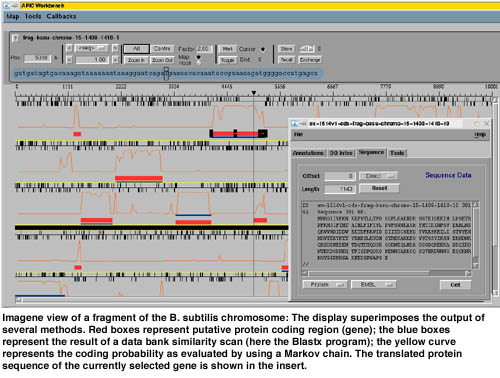
Imagene is the result of a five years collaboration between INRIA, Université Paris 6, the Institut Pasteur and the ILOG company. The system has been implemented by using an object oriented model and a co-operative solving engine provided by ILOG. In Imagene, a global problem (task) is solved by successive decompositions into smaller sub-tasks. During the execution, the various sub-tasks are graphically displayed to the user. In that sense, Imagene is more transparent to the user than a traditional menu-driven package for sequence analysis since all the steps in the resolution are clearly identified. Moreover, once a task has been solved, the user can restart it at any point; the system then keeps track of the different versions of the execution. This allows to maintain several hypothesis in parallel during the analysis. Imagene also provides a user interface to display, on the same picture, the results produced by one or several strategies (see Figure). Due to the homogeneity of the whole software, this display is fully interactive and the graphical objects are directly connected to their database counterpart.

Imagene has been used within several bacterial genome sequencing projects (Bacillus subtilis and Mycoplasma pulmonis) and has proved to be particularly useful to pinpoint sequencing errors and atypical genes. However this first version suffers several drawbacks. First it was limited to the representation of prokaryotic data only, second the development tools were commercial thus giving rise to difficulties in its diffusion, last, it was designed to handle pure sequence data from a single genome. In order to overcome these limitations, we undertook a new project (Geno-Annot) through a collaboration between INRIA, the Institut Pasteur and the Genome-Express biotech compagny. As a first step, the data model was extended to eukaryotes and completely re- implemented using the AROM system developed at INRIA (http://www.inrialpes.fr/romans/pub/arom). We are now in the process of re-designing the task-engine and the graphical user interfaces in JAVA. Finally, our ultimate goal will be to integrate Geno-Annot within a more general environment (called Geno-*) in order to fully link all the pieces of genomic information together (ie sequence data, metabolism, gene expression etc). Geno-Annot is a two years project that started in September 1999.
Links:
Action Helix: http://www.inrialpes.fr/helix.html
Imagene: http://abraxa.snv.jussieu.fr/imagene
Please contact:
Alain Viari - INRIA
Tel: +33 4 76 61 54 74
E-mail: alain.viari@inrialpes.fr