

ERCIM News No.43 - October 2000 [contents]
Configurable DNA Computing
by John McCaskill
The DNA Computing Project carried out at the GMD Biomip Institute, aims at making molecular systems more programmable. Computer scientists, chemists, molecular biologists, physicists and microsystem engineers are working together to produce both a technological platform and theoretical framework for feasible and evolvable molecular computation.
Although the massive parallelism of DNA in solution is impressive (more than 1020 bytes of active memory per liter) and the energy consumption is very low, the ultimate attraction of DNA-Computers is their potential to design new hardware solutions to problems. Unlike conventional computers, DNA computers can construct new hardware during operation. Thus, the closest point of contact to electronic computing involves hardware design, in particular reconfigurable hardware design, rather than conventional parallel algorithms or languages. Molecular computers can be constructed reversibly in flow systems, where an exchange of DNA populations is possible. Rapid hardware redesign opens the door to evolving computer systems, so that configurable DNA Computing also aims at harnessing evolution for design and problem solving. Because of the huge information storage potential of aqueous solutions containing DNA, comparatively low flow rates suffice for massively parallel processing so that synthetic DNA can be treated as an affordable, easily degradable resource. Sequence complexity on the other hand is expensive to purchase but is generated within the DNA Computer, starting from simple sequence modules.
The GMD DNA Computing Project is multidisciplinary in scope, aiming at making molecular systems more programmable. Computer scientists, chemists, molecular biologists, physicists and microsystem engineers are working together to produce both a technological platform and theoretical framework for an effective use of molecular computation. The initial barrier is the issue of complexity: just how scalable and programmable are the basic hybridisation processes underlying DNA Computing? The group has devised an optically programmable, scalable concept for DNA Computing in microflow reactors, designed to be extensible to allow full evolutionary search (see below). No flow switching is required (since this is not currently scalable) in this dataflow like architecture. Currently the microreactor geometry is fixed to evaluate scalability on a benchmark problem: Maximum Clique. Specific problem instances are reconfigured optically.
 |

|
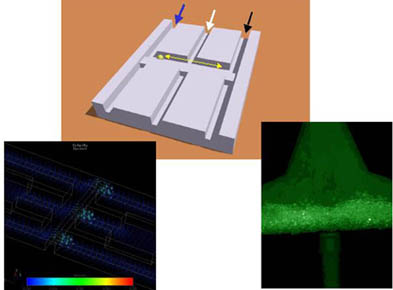
| Figure 1: Massively parallel subset selection module for DNA Computing: The three images show a schematic, hydrodynamic simulation and fluorescence image of a microstructured selection module made at the GMD. The concept is that a subset of a mixed population of DNA flowing through the left hand side of the reactor binds to complementary DNA sequence labels attached to magnetic beads. When the beads are transferred (synchronously for all such modules) to the right hand side, they enter a denaturing solution which causes the bound DNA subset to be released. The released DNA subset solution is neutralised before being subjected to further modules. The hydrodynamic flow in one such module is seen in the color coded image on the left. Continuous flow preserves the integrity of the two different chemical environments in close proximity in each module. The fluorescence image shows DNA hybridized to the DNA labels on magnetic beads in such a microreactor, allowing the DNA processing to be monitored. |
Figure 2: Medium Scaleup Programmable Microflow System for Benchmark Problem: Portion of the two layer mask design structure for integrating many selection modules (see Fig. 1) to solve a combinatorial optimization problem - in this case maximal clique. The structures are etched into a silicon substrate on the top and bottom sides (green and red), with through connections at the sites of squares. The microflow reactor is sealed at top and bottom with anodically bonded pyrex wafers and fitted with connecting tubing as shown in Fig. 3. This particular microreactor design can solve any intermediate scaled instance of the clique problem up to N=20. Which instance is programmed optically by directing the attachment of DNA to beads. |
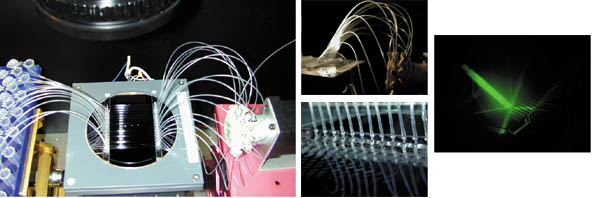 |
| Figure 3: Experimental DNA Computing Apparatus: The left photo shows a microreactor with attached tubing connected to a multivalve port on the right and liquid handling system (not shown). The microreactor is imaged under green laser light to detect fluorescence at different locations stemming from DNA molecules, monitoring the time course of the computation. A microscope setup, not shown, is also used for reading and writing. The images on the right show close ups of the fluidic connections to the microreactor and of the laser illumination of the wafer. |
Effective DNA Computing is dependent on the construction of a powerful interface to the molecular world. In this project, the interface involves configurable microreactors with photochemical input and fluorescence readout down to the single molecule level. Fluorescence detection is the most sensitive spectroscopic technique for detecting the presence of specific molecules in solution and it can be employed as an imaging tool to gain vast amounts of information in parallel about the status of computations or the final answer. Consistent with our perspective on DNA Computing as hardware design, we employ photolithographic techniques to program the attachment of DNA to specific mobile elements within the microreactor. At the spatial resolution and time scale required, photolithographic projection can be made completely dynamically programmable.
Directed molecular evolution provides a second stepping stone to exploiting more powerful algorithms in DNA Computing. The programming problem shifts to defining selection conditions which match the given problem. The strategy employed in this project is to program molecular survival by employing flow networks involving sequence-dependent molecular transfers in series and parallel. Feedback loops and amplification modules have been designed and will be introduced into the computer in due course to complete the integration with molecular evolution. Reconfiguration and evolution of the compartmentation and flow network is planned at a second phase in the technology development to increase the general programmability of the computer.
DNA-Computers can produce their calculated output in functional molecular form for direct use. Application areas include the pharmaceutical and diagnostics industry (where molecular complexity requires increasingly sophisticated algorithms for combinatorial libraries construction and readout implemented in molecular hardware), nanotechnology and high performance computing for coding, associative retrieval and combinatorial optimisation.
Status of the Project
A scalable architecture for configurable DNA Computing has been developed for fully optically programmable solution of the NP-Complete test problem "Maximal Clique". Individual modules (strand transfer and amplification) have been implemented in microreactors and the optical programmability has been developed and demonstrated. Spatially resolved single molecule fluorescence detection has been developed to allow an on-line readout of information in complex DNA populations. Microsystem integration has proceeded up to the level of 20x20x3 selection modules, but current evaluation work is of a 6x6x3 DNA microprocessor. A DNA library for medium scale combinatorial problems (N=32) has been designed and the lower portion of it constructed. The potential of iterated modular evolution in DNA Computing has been evaluated on a massively parallel reconfigurable electronic computer (NGEN). All of these developments have alternative technological applications beyond the immediate range of DNA Computing.
Cooperation
Collaborations are being fostered with the European Molecular Computing Consortium (EMCC), in particular University of Leiden, Holland (Prof. G. Rozenberg, Prof. H. Spaink); North Rhine Westfalia initiative in Programmable Molecular Systems: including University of Cologne (Prof. Howard), University Dortmund (Prof. Banzhaf) and University Bochum (Prof. Kiedrowski).
Links:
BioMIP website: http://www.gmd.de/BIOMIP
Please contact:
John S. McCaskill - GMD
Tel: +49 2241 14 1526
E-mail: McCaskill@gmd.de



