
by Andy S. Law and Alan L. Archibald
The Bioinformatics Group at the Roslin Institute develops tools and resources for farm animal genome analysis. These encompass the databases, analytical and display tools required for mapping complex genomes. The World Wide Web is used to deliver the resources to users.
The Bioinformatics Group at the Roslin Institute aims to provide access to appropriate bioinformatics tools and resources for farm animal genome analysis. Genome research in farm animals is largely concerned with mapping genes that influence economically important traits. As yet, there are no large-scale genome sequencing activities. The requirements are for systems to support genetic (linkage), quantitative trait locus (QTL), radiation hybrid and physical mapping and to allow data sharing between research groups distributed world-wide.
resSpecies – a Resource for Linkage and QTL Mapping
Genetic linkage maps are constructed by following the co-segregation of marker alleles through multi-generation pedigrees. In quantitative trait locus (QTL) mapping, the performance of the animals is also recorded. Both QTL and linkage mapping studies require databases to store and share the experimental observations. Data sharing between research groups is particularly valuable in linkage mapping. Only by pooling data from the collaborating groups can comprehensive maps be built.
We developed resSpecies to meet this need. It uses a relational database management system (RDBMS - INGRES) with a web-based interface implemented using Perl and Webintool (Hu et al. 1996. WebinTool: A generic Web to database interface building tool. Proceedings of the 7th International Conference and Workshop on Database and Expert Systems (DEXA 96), Zurich, September 9-13, 1996 pp 285-290). This makes international collaborations simple to effect.
The relational design ensures that complicated pedigrees can be represented relatively simply. Populations are defined as groups of individuals. Within resSpecies, access is granted to individual contributors/collaborators on each population separately. The database stores details of markers and alleles. Genotypes may be submitted through a simple web interface that infers missing genotypes, checks for Mendelian inheritance and rejects data that contains inheritance errors. Using a series of simple query forms, data can be extracted in the correct format expected by a number of popular genetic analysis algorithms (eg crimap). This eliminates the possibility of cryptic typographical errors occurring and ensures that the most up-to-date data is available at all times.
resSpecies is used to support Roslin’s internal programmes and several international collaborative linkage and QTL mapping projects.
ARKdb – a Generic Genome Database
Scientists engaged in genome mapping research also need access to contemporary summaries of maps and other genome-related data.
We have developed a relational (INGRES) genome database model (ARKdb) to handle these data, along with web-based tools for data entry and display. The information stored in the ARKdb databases includes linkage and cytogenetic map assignments, polymorphic marker details, PCR primers, and two point linkage data. Each observation is attributed to a reference source. Hot links are provided to other data sources eg sequence databases and Medline (Pubmed).
The ARKdb database model has been implemented for data from pigs, chickens, sheep, cattle, horses, deer, turkeys, cats, salmon and tilapia. The full cluster of ARKdb databases are mounted on the genome server at Roslin with subsets at Texas A+M and Iowa State Universities. We have also developed The Comparative Animal Genome database (TCAGdb) to capture evidence that specific pairs of genes are homologous. We are developing automated Artificial Intelligence methods to evaluate homology data.

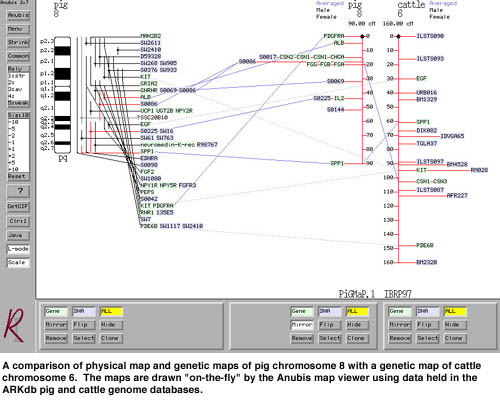
The Anubis Map Viewer
Visualisation is the key to understanding complex data and tools that transform raw data into graphical displays are invaluable. The Anubis map viewer was the first genome browser to be operable as a fully-fledged GUI (Graphical User Interface) over the Web. It is used as the map viewer for ARKdb databases and the INRA BOVMAP database. We have recently launched a prototype java version of Anubis - Anubis4.
Future Activities
We are developing systems to handle the data from radiation hybrid, physical (contig) mapping, expression profiling (microarray) and expressed sequence tag (EST) experiments. Exploitation of the wealth of information from the genomes of human and model organism is critical to farm animal genome research. Therefore, we are exploring ways of improving the links and interoperability with other information systems. Our current tools and resources primarily address the requirements for data storage, retrieval and display. In the future we need to fully integrate analytical tools with the databases and display tools.
The Roslin Bioinformatics Group has grown to eleven including software developers, programmers and database curators. In the past we have received support from the European Commission and Medical Research Council. The group is currently funded by grants from the UK’s Biotechnology and Biological Sciences Research Council.
Links:
http://www.roslin.ac.uk/bioinformatics/
Please contact:
Alan L. Archibald - Roslin Institute
Tel: +44 131 527 4200
E-mail: alan.archibald@bbsrc.ac.uk