Combating Illiteracy in Chemistry: Towards Computer-Based Chemical Structure Reconstruction
by Marc Zimmermann, Le Thuy Bui Thi and Martin Hofmann
The majority of chemical structure information in the literature (including patents) is present as two-dimensional graphical representations. Although these chemical structures have usually been produced with a chemical drawing program, the machine-readable compound information is lost in the course of publication. Currently, 90% of published structures are not in a chemical file format that can be interpreted by a computer. At the Fraunhofer Institute for Algorithms and Scientific Computing, a project has been initiated to extract chemical information from depictions of molecules in the public literature. The goal of this work is to gather the rich information from pharmaceutical patents and transform it into a machine-readable format.
|
Our research group in the Department of Bioinformatics at Fraunhofer SCAI is working on the automated extraction of information from biomedical literature. In this highly interdisciplinary domain, interesting information is often presented as a combination of text and graphics. Based on our experience in the field of biological information extraction (eg protein-protein interaction networks), we recently extended the scope of our research towards chemical entity recognition and chemical structure reconstruction.
The information described in patents is extremely valuable to researchers involved in the design of potential drugs. A patent comprises information on the complete drug-design process, including the disease and indication area, the target protein, the chemical structure of the drug molecules and assay information. Chemical information is often available in printed form or as bitmaps in on-line resources. Reproducing this information by redrawing the structure with a computer program is time-consuming and prone to errors. This process could be greatly improved by a system that was able to read and understand chemical drawings, and could automatically extract the information necessary for a public database.
State-of-the-Art
While the problem of chemical structure recognition and reconstruction is not new, it has not yet been solved. The first attempt at chemical structure recognition resulted in CLiDE (Chemical Literature Data Extraction, developed by the University of Leeds in the beginning of the nineties). We have tested CLiDE in an extensive validation study with drawings of pharma-relevant molecules. The results of the study are based on an assembled test-set of 100 'blockbuster' drug (best-selling drugs, see www.rxlist.com/02top.htm) from 2002. The structures were extracted from the Merck Index and were also manually drawn using Chemdraw (a widely used drawing tool for molecule structures). These 200 files were submitted to CLiDE and the output was manually inspected. Almost 50% had at least one error and often the recognition process was interrupted and asked for manual interference (on average twice per molecule). Unfortunately, there seems to be no further public or commercial effort to improve CLiDE (eg enabling the tool to learn from human intervention).
|
Scientific Challenge
Following our validation study, we have taken up the challenge to develop a prototype for a new structure recognition tool that would enable true chemical structure reconstruction. In order to overcome the shortcomings of CLiDE, we propose to link modern supervised machine-learning algorithms (eg Support Vector Machines) with cheminformatics similarity searching and reaction-planning algorithms. The following scientific challenges will be addressed:
- conversion of the raster image format into vector graphics
- elimination of 'graphic noise' through a filter module
- development of a graph grammar to handle contours, additional or missing lines or nodes and intersections
- matching of small sub-graphs to a database of consensual chemical fragments
- merging of identified sub-graphs into a single molecular graph based on chemical synthesis rules (ie a chemical grammar)
- training and machine intelligence.
First Steps
For this final challenge, a rule-based approach to the mapping of overlapping fragments from the graph matcher can increase the reliability of the reconstructed molecule. The filter rules, the collection of common fragments and the synthesis rules will initially be trained on a test corpus. Afterwards the whole process can be further improved by automatically retraining each module with the output of the manual curation tool.
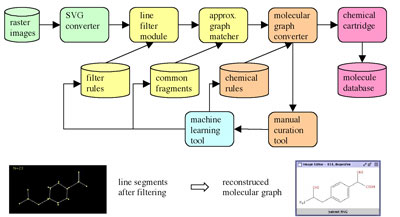
In order to solve the problem of recognizing and learning chemical structures in image documents, our system combines pattern recognition techniques with supervised machine-learning concepts. The method is based on the idea of identifying from depictions the most significant fragments of small molecules. We have therefore recursively decomposed a diverse collection of 'blockbuster' molecules into smaller sub-graphs in an off-line process. At runtime, the input image of a chemical structure is converted into a vector graphic. A graph representation of this vector graphic is then generated by combining line segments. In this pre-processing step, the trained system detects and corrects any common errors that occurred during the vectorization procedure or were due to a flawed scanned image. The next step uses sub-graph isomorphism to detect known fragments in the assembled graph. Finally all sub-graphs are combined using a chemical knowledge image (ie proposing chemically suitable corrections) in order to reconstruct the most likely structure represented in the input. If the input graph cannot be matched completely, the user will be asked to specify the solution with a manual curation tool; the solution is then added to the knowledge space of trained molecules. In this way we expect the system to steadily improve its performance.
We have already assembled a test corpus of blockbuster molecules and their fragments. We have integrated potrace (http://potrace.sourceforge.net) and autotrace (http://autotrace.sourceforge.net) as SVG converter tools, and the pre-processing and the graph-matching module have been implemented. The next steps are to separate atom labels from chemical bonding lines in the input image and to recognize them. The final step is then to convert the reconstructed graph to a chemical file format.
Please contact:
Marc Zimmermann, Fraunhofer Institute for Algorithms and Scientific Computing SCAI,
Fraunhofer ICT Group, Germany
E-mail: marc.zimmermann![]() scai.fraunhofer.de
scai.fraunhofer.de

 This issue in
This issue in