The Prognochip Project: Transcripromics and Biomedical Informatics for the Classification and Prognosis of Breast Cancer
by Dimitris Kafetzopoulos
The completion of the Human Genome Project and the development of post-genomic applications have allowed new holistic approaches to disease analysis that will revolutionize biomedical research and health care. Consultation of both the comprehensive genotypic information of the patient and the detailed molecular classification of the disease will result in individualized treatments. The Greek 'Prognochip' project applies this approach in the field of breast cancer prognosis and treatment.
Breast cancer affects approximately one in ten women, and as such is one of the most common female malignancies. Breast cancer is both genetically and histo-pathologically heterogeneous, and the mechanisms underlying its development remain largely unknown. Breast cancer patients diagnosed with the same stage of disease often have remarkably different responses to therapy. Even with the strongest prognostic indicators, such as lymph-node status, estrogen receptor expression and histological grade, it is not possible to accurately classify breast tumours according to their clinical behavior. Genomic background and variations in the transcriptional programs account for much of the observed diversity. The 'Prognochip' project, funded by the Greek General Secretariat for Research and Technology, aims to identify and validate 'signature' gene expression profiles of breast tumours that correlate with other epidemiological or clinical parameters. This should provide a more accurate prognosis and prediction of response to therapy: a clear benefit to almost three out of four women who receive aggressive chemotherapy treatment, although they would have survived without it.
The major tasks within the Prognochip project are as follows:
Patients are informed and consent to the molecular and genetic data analysis of their tumour specimens and blood samples, provided that their anonymity is ensured. A tissue procurement protocol has been designed for tissue collection and storage and a tissue-bank system has been established for proper tissue filing and management. Patients with malignant tumours are staged according to the TNM system and a set of immunohistological markers are examined. A DNA microarray of long oligonucletide probes has been designed, representing all known human genes – approximately 35,000 different transcripts of 27,000 different genes. A common reference material has been chosen for the study, consisting of a defined set of cell-line extracts, thereby ensuring accurate quantitation of gene expression. After hybridization, fluorescence intensity images representing gene expression levels are acquired as 16-bit TIFF files and stored in BASE, a MySQL database developed by the University of Lund, Sweden. Special plug-ins have been created for data pre-processing (filtering, normalization) and analysis.
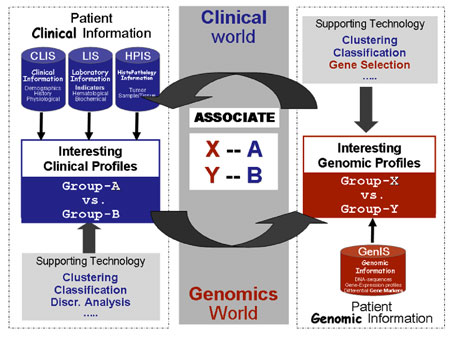
In general, two computational approaches from a suite of intelligent data processing tools are used for tumour classification. The first approach is the 'unsupervised' analysis, in which no source of knowledge is used to guide the analysis process. Instead, the data are searched for patterns with no a priori expectation concerning the number or type of groups (gene and tumour clusters) that might be present. The second is the 'supervised' analysis, in which we search for genes whose expression patterns (denoted X and Y in Figure 1) correlate with external parameters (denoted A and B). The 'supervising' parameters are clinical features such as the clinical outcome (including overall survival, relapse-free survival times, metastasis etc), other molecular markers, chromosomal aberrations, patterns observed with other diagnostic methods and responses to (chemo)therapy.
|
In addressing classification, there are two issues: a) class discovery, the definition of previously unrecognized tissue sub-types, and b) class prediction, the assignment of particular samples to existing classes (this could reflect current states or even future outcomes).
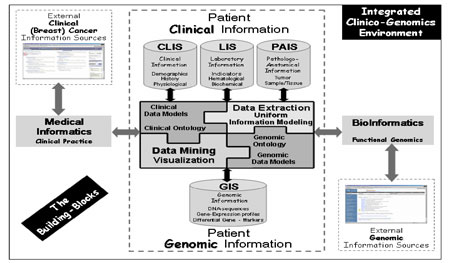
The main challenge of the Prognochip prospective study is the development of interoperable and efficient clinical and genomic information systems for the integration of heterogeneous data. In that context we are working towards the delivery of an Integrated Clinico-Genomics Information Technology Environment (ICG-ITE). The envisioned building blocks of the ICG-ITE include:
- a set of clinical information systems to store patients' clinical information
- an information system to store and manage the specifications of the respective microarray experiments, analyse measured bioassaysand store samples' genomic information
- a middleware layer for information/ data integration and intelligent processing.
The middleware layer is realized through a 'puzzle' of integrated software components that together enable:
- the seamless and efficient extraction of data from various sources (clinical and genomic)
- uniform information modelling through the utilization of standard clinical/genomic data models and respective ontologies
- uniform information representation through the utilization and appropriate customization of RDF/XML technology
- intelligent data processing and visualization through a suite of data mining components and tools.
|
Since the integration of clinical and genomic data is such a demanding problem, there is a clear need to elaborate on the concept of Integrated Electronic Health Care Record architectures, utilize technological advances and extend the standard clinical data models to include and amalgamate genomic ones. Further, an equally important security and authorization infrastructure is employed. A general layout of the provisioned ICG-ITE is shown in Figure 2.
Please contact:
Dimitris Kafetzopoulos, IMBB-FORTH
Tel: +30 2810 39 1594
E-mail: kafetzo![]() imbb.forth.gr
imbb.forth.gr

 This issue in
This issue in