Spatio-Temporal Analysis in 4D Video-Microscopy
by Charles Kervrann, Jérôme Boulanger and Patrick Bouthemy
Recent progresses in cellular, molecular biology and light microscopy make possible the acquisition of multidimensional data (3D+time) and the observation of dynamics of fast cellular activities. Image processing methods for quantitative analysis of these massive movements have been limited. Automatic techniques to extract information about dynamics from image sequences are therefore of major interest, for instance, to assess the role of Rab GTPases, a large gene family, involved in membrane transport. We have recently developed methods to perform the computational analysis of 4D image sequences.
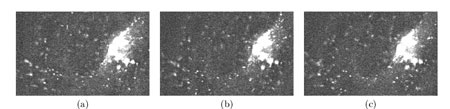
This project aims at extracting major motion components by statistical learning and spatial statistics from the computed partial or complete trajectories. We mainly focus on the analysis of vesicles that deliver cellular components to appropriate places within cells. Applications of the proposed image processing methods to biological questions should provide a new and quantitative way for interpreting motility of membrane transport vesicles. The challenge is to track Green Fluorescent Protein (GFP) tags with high precision in movies representing several gigabytes of image data and collected and processed automatically to generate information on complex trajectories. Quantitative analysis of data obtained by fast 4D deconvolution microscopy allows to enlighten the role of specific Rab proteins. The role of Rab proteins is viewed as to organize membrane platforms serving for protein complexes to act at the required site within the cell. Methods have been developed for a target protein - Rab6a' - involved in the regulation of transport from the Golgi apparatus to the endoplasmic reticulum. Typically, the state of Golgi membranes during mitosis is controversial, and the role of Golgi-intersecting traffic in Golgi inheritance is unclear. In Figure 1, three types of objects are observed within the cell: 1) organelle (Golgi), 2) rapid transport intermediates (moving vesicles) and 3) elongated structures (membrane tubulations).
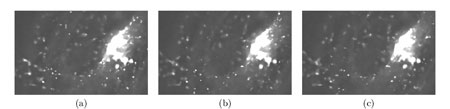
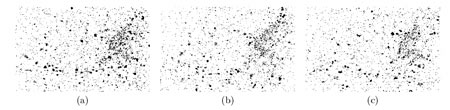
Work began with an analysis of possible methods to improve the quality of images and their adaptation to 4D imaging. We present a spatio-temporal filtering method for significantly increasing the signal-to-noise ratio in noisy fluorescence microscopic image sequences where small particles have to be tracked from frame to frame. New video-microscopy technologies allow to acquire 4-D data that require the development and implementation of specific image processing methods to preserve details and discontinuities in both the three x-y-z spatial dimensions and the time dimension. Particles motion in such noisy image sequences cannot be reliably calculated since objects are small and untextured with variable velocities; the S/R ratio is also quite low due to the relatively limited amount of light. However, partial trajectories of objects are line-like structures in the spatio-temporal x-y-z-t domain. Image restoration can be then achieved by an adaptive window approach which has been already used to efficiently remove noise in still images and to preserve spatial discontinuities, for image decomposition into noise, texture and piecewise smooth components. The proposed 'adaptive window approach' is conceptually very simple being based on the key idea of estimating a locally regression function with an adaptive choice of the space-time window size (neighbourhood) for which the applied model fits the data well. We use statistical 4-D data-driven criteria for automatically choosing the size of the adaptive growing neighbourhood. At each pixel, we estimate the regression function by iteratively growing a space-time window and adaptively weighting input data to achieve an optimal compromise between the bias and variance. The proposed algorithm complexity is actually controlled by simply restricting the size of the larger window and setting the window growing factor. Global statistics such as velocities and directions could be then extracted in order to measure the general behaviour of vesicles in the 4D volume.
|
We have applied this method to noisy synthetic and real 4-D images where a large number of small fluorescently-labeled vesicles move in regions close to the Golgi apparatus. The S/R ratio is shown to be drastically improved resulting enhanced objects which even can be segmented. The objective is to report evidences about the lifetime kinetics of specific Rabs in different membranes may be similar within on cell type. This novel approach can be further used for biological studies where dynamics have to be analyzed in molecular and subcellular bioimaging.
This work was performed in collaboration with the MIA Unit (Mathématiques et Informatique Appliquées) from INRA, Jouy-en-Josas, Curie Institute - UMR 144 - CNRS ('Compartimentation et Dynamique Cellulaires' Laboratory), Paris and UMR 6026 ('Interactions Cellulaires et Moléculaires' Laboratory – 'Structure et Dynamique des Macromolécules' team), Rennes. The Vista team is the prime contractor of this project (MODYNCELL5D).
A second applicative project will concern the CLIP 170 protein involved in the kinetochores anchorage (in the segregation of chromosomes to daughter cells, the chromosomes appear to be pulled via a so-called kinetochore attached to chromosome centromeres) using new fluorescent probes (Quantum Dots). New image analysis methods should be developed for tracking fluorescent molecules linked to microtubules.
Please contact:
Charles Kervrann, IRISA/INRIA, France
Tel: +33 2 99 84 22 21
E-mail: ckervran![]() irisa.fr
irisa.fr

 This issue in
This issue in