myGrid: Middleware for In Silico Experiments in Biology
by Carole Goble
Life science researchers traditionally chain together database searches and analytical tools, using complex scripts to overcome incompatibilities, or by manually cutting and pasting between web interfaces. These 'in silico' experiments are usually undertaken without support for the scientific process of managing, sharing and reusing the results, their provenance, and the methods used to generate them. The myGrid project has developed a comprehensive loosely-coupled suite of middleware components specifically to support data intensive in silico experiments in biology. Workflows and query specifications link together third party and local resources using web service protocols. The software can be freely downloaded and has been used for building discovery workflows by Williams-Beuren Syndrome and Grave's Disease Life Scientists collaborating with myGrid.
Bioinformaticians are knowledge workers, intelligently weaving together information globally available to the community, linking and correlating it meaningfully, and generating even more information. However, they are commonly forced to waste their time overcoming incompatible interfaces to applications and neglect basic scientific practice such as keeping records of the methods they used to integrate applications, how results link together, and even the goal of the experiment. The omission of these provenance records makes in silico experiments difficult to interpret and hard to repeat, and the methods difficult to reuse. This is not because bioinformaticians are unskilled but because the appropriate infrastructure to support them in their tasks are missing. The myGrid project has researched and developed open source high-level service-based middleware to support in silico experiments in biology, using workflows and semantic technologies.
The project, which has been running since late 2001, is a UK EPSRC-funded e-Science pilot project made up of a consortium of the Universities of Manchester, Newcastle, Nottingham, Sheffield and Southampton, IT Innovation, and the European Bioinformatics Institute, supported by nine industrial partners of whom GSK and IBM are the most significant. A third of the team are bioinformaticians with a Life Science background. The aim is to develop generic middleware driven by the real day to day problems of bioinformaticians, so the project has been firmly guided by strategic partnerships with UK-based Life Scientists in Newcastle-upon-Tyne investigating Grave's Disease, in Manchester investigating Williams-Beuren Syndrome and, more recently, in Liverpool investigating Trypanosomiasis in cattle. Third party developers of the EMBOSS, BioMOBY and SeqHound bio-service suites also use the middleware as a means of accessing and integrating their services. Currently myGrid supports access to over 600 public services.
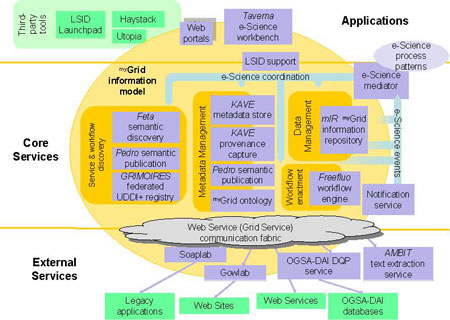
The middleware is a toolkit of core components for forming, executing, managing and sharing discovery experiments. The components are intended to be adopted in a 'pick and mix' way by developers and tool builders to produce end applications. Bioinformaticians and service providers develop and run experiments via the Taverna workbench whereas biologists may use our configurable web portal to run them, examine results, and collaborate. Workflows that execute remote or local web services and Java applications are the chief mechanism for forming experiments. Legacy applications are incorporated using our Soaplab-Gowlab wrapper tools. Any web service can be incorporated – there is no restriction on the type of biology. The Taverna workbench is a GUI used for assembling, adapting and running workflows enacted by the Freefluo workflow enactment engine (see Figure 1). The project's Scufl workflow language is a user-oriented abstraction over general graph languages hiding the details of service invocation and control flow. Users can also configure support for fault management and service failover. In addition to workflows, databases may be integrated using the OGSA-Distributed Query Processor developed jointly with the UK OGSA-DAI project.
|
To support the scientific method the project has adopted a number of innovative technologies from the semantic web community. To enable service providers and bioinformaticians to publish, discover and match-make services, and bioinformaticians to publish and reuse workflows, our registry has mechanisms to support descriptions drawn from an ontology defined in the W3C standards RDF (Resources Description Framework) and OWL (Web Ontology Language). Our Knowledge Annotation and Verification of Experiments component (KAVE) captures and stores provenance records of methods and purpose in RDF, and again semantically annotated by terms from an ontology.
The consortium has followed a twin-track of core-development and research-prototyping. Research efforts in semantic services and metadata management are currently being migrated to production as part of the core development. Through the use of standards such as Web services, RDF, OWL and Life Science Identifiers (LSIDs), the project has been able to leverage third party tools such as IBM's Haystack and LSID LaunchPad.
As the current funding finishes, many challenges remain. myGrid's ability to rapidly create and run in silico experiments is now being used by a range of newly funded Life Science projects such as PsyGrid, CLEF-Services, e-Fungi, ISPIDER, Integrative Biology and ComparaGrid. These and other projects, such as the EU FP6 STREP OntoGrid, also contribute to our continued middleware development. Further technical challenges in integrating disparate and distributed applications include: the incorporation of interactive, computationally intensive simulation services, such as heart models; the efficient management of very large data sets; the visualisation of results; and knowledge mining provenance metadata.
Links:
myGrid: http://www.mygrid.org.uk
OGSA-DAI: http://www.ogsadai.org.uk
Please contact:
Carole Goble,
The University of Manchester,UK
E-Mail: carole.goble@manchester.ac.uk

 This issue in
This issue in