| |
Virtual Vascular Surgery on the Grid
by Peter Sloot and Alfons Hoekstra
Medical simulations and visualizations typically require computational power not usually available in a hospital. The University of Amsterdam recently demonstrated Virtual Vascular Surgery (a virtual bypass operation), where large-scale simulation and visualization capabilities were offered as services on the Grid.
Arteriosclerosis is a widespread disease that manifests particularly in developed countries. Treatment often involves surgery, such as the placement of bypasses that lead the blood around clogged arteries to restore normal blood flow. A surgeon plans these interventions on the basis of 3D images obtained from MRI or CT scans. Apart from considerations such as accessibility, the attainable improvement in the blood flow will determine what type of treatment is appropriate. Improvements in the support for planning these procedures are expected to improve their success rate. We have developed a prototype grid-based system for virtual vascular surgery, which may be used during pre-operative planning or as a valuable tool in the training of novice vascular surgeons. The prototype uses advanced distributed simulations and visualizations to support vascular surgeons in making pre-operative decisions.
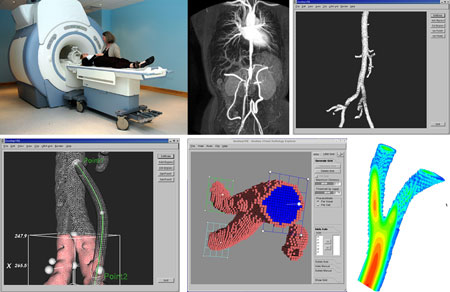 |
| Figure 1: Distributed image-based blood-flow simulation and visualization on the Grid. From top left to bottom right: A patient is scanned in Leiden, The Netherlands; this results in a raw image stored in a Storage Element on the Grid (eg Poznan, Poland); this is segmented, filtered and cropped, using a Grid service; a bypass is added; a computational mesh is generated (on the local machine) and given to the parallel flow solver, running on Compute Elements in the Grid (Amsterdam and Spain); the resulting flow fields are displayed on the local machine using visualization services offered by the Grid Visualization Kernel (Linz, Austria). |
The Virtual Radiology Explorer
The Virtual Radiology Explorer (VRE), developed at the University of Amsterdam, is a grid-based problem-solving environment for virtual vascular surgery. The VRE contains an efficient mesoscopic computational haemodynamics solver for blood-flow simulations based on parallel cellular automata. To convert the medical scans into computational meshes, raw scanner data is first segmented so that only the arterial structures of interest remain in the data set. The segmented data is then converted into a computational mesh. The patient’s blood flow is simulated using grid resources. The VRE system uses a desktop virtual reality environment where the patient’s data, obtained from a scanner, is visualized as a 3D stereoscopic image together with the graphical interpretation of the simulation results. A user can then manipulate the 3D images of the arteries. This would be the virtual surgical procedure, eg the addition of a bypass. Blood flow in this new geometry is also computed, and the user can then assess the effectiveness of the proposed treatment, and might try other alternatives to optimise the procedure.
The medical scanners, the visualization environment and the computational resources required for the flow computations and visualizations are usually located in disparate geographical locations and distinct administrative domains. The need for transparent access to these resources, high efficiency and strict data security triggered the development of our advanced grid-based simulation and interactive visualization for virtual vascular surgery. The middleware to support this was developed in the CrossGrid project.
CrossGrid
The CrossGrid project is oriented towards compute- and data-intensive applications characterized by the interaction with a person in the loop. The CrossGrid pan-European distributed testbed shares resources across sixteen European sites. One key component of Crossgrid is the Migrating Desktop (MD) grid portal. The MD produces a transparent user work environment, permitting the user to access grid resources from remote computers. Users can run applications, manage data files, and store personal settings independently of the location or the terminal type. We have incorporated our VRE system into the Grid via the MD grid portal. We achieved secured grid access, node discovery and registration, grid data transfer, application initialization, medical data segmentation, segmented data visualization, computational mesh creation, job submission, distributed blood-flow visualization, and bypass creation. VRE runs on a local machine but is launched and initialized through the MD. The input takes the form of segmented or non-segmented medical data produced at the Leiden Medical Centre (LUMC); the CrossGrid testbed provides access to this data from a medical image repository acting as a Grid Storage Element in Leiden.
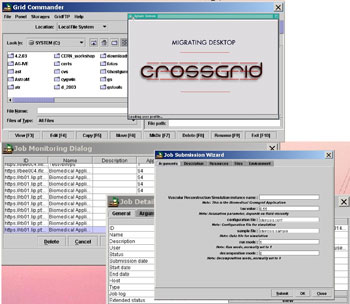 |
| Figure 2: The CrossGrid Grid Portal – the ‘migrating desktop’ – with some of its functionalities, eg grid log-in and Grid Proxy creation, virtual node navigation and Grid data transfer (via the Grid Commander), submission of blood flow simulations to the CrossGrid testbed (via the Job Submission Wizard) and monitoring of jobs running in the testbed (via the Job Monitoring Dialog). |
A Virtual Bypass Operation on the Grid
We have recently demonstrated the following scenario: The abdominal aorta area of a patient is scanned, and the resulting image is stored in a Radiology Information System repository. Later, a physician (user) logs into the CrossGrid Portal using his Grid certificate and private key. The user checks if there are segmented or non-segmented medical data ready for analysis in one of the virtual nodes, and securely transfers a few to his local machine. The user then starts the VRE from within the MD, loads the segmented medical data, selects a region of interest, crops the image, adds a bypass, and creates a computational mesh. The user selects the Biomedical Application icon within the MD (with parameters and files being taken from the user's profile), and submits the job to the CrossGrid testbed, to the nearest or most adequate Computing Element in the Grid. The user may then check job submission or progress via the MD. After the job has been completed, the calculated velocities, pressure, and shear stress are transferred to the local Storage Element or to the Grid Visualization Kernel to be rendered and reviewed by the user. We will give a live demonstration of this process during the upcoming European Grid Conference in Amsterdam, February 2005.
Overall, the successful deployment of grids requires the re-design of algorithms to support loosely coupled computer resources. This should be driven by relevant and challenging applications. With the experience gained within CrossGrid from the virtual vascular surgery application (and three other applications), European Grid Technology leads the way in new Grid developments worldwide.
Links:
http://www.science.uva.nl/research/scs
http://www.crossgrid.org/
Please contact:
Peter Sloot, University of Amsterdam,
The Netherlands
Tel: +31 20 525 7462
E-Mail: sloot science.uva.nl science.uva.nl
|




