
by Niko Beerenwinkel, Joachim Selbig, Rolf Kaiser and Daniel Hoffmann
To develop tools that assist medical practitioners in finding an optimal therapy for HIV-infected patients – this is the aim of a collaboration funded by Deutsche Forschungsgemeinschaft that has been started this year by researchers at GMD, the University of Cologne, CAESAR, the Center of Advanced European Studies and Research, Bonn, and a number of cooperating university hospitals in Germany.
The Human Immunodeficiency Virus (HIV) causes the Acquired Immunodeficiency Syndrome (AIDS). Currently, there are two types of drugs in the fight against HIV, namely inhibitors of the two viral enzymes protease (PR) and reverse transcriptase (RT).
Since HIV shows a very high genomic variability, even under the usual combination therapy (HAART – highly active antiretroviral therapy) consisting of several drugs, mutations occur, that confer resistance to the prescribed drugs and even to drugs not yet prescribed (cross resistance). Therefore, the treating physician is faced with the problem of finding a new therapy rather frequently.
Clinical trials have shown that therapy changes based on a genotypic resistance test, ie sequencing of the viral gene coding for PR and RT and looking for mutations known to cause resistance, result in a significantly better therapy success. However, not all patients benefit from therapy changes after resistance testing. There are several possible reasons for therapy failure in this situation: the occurrence of an HIV-strain resistant to all available antiretroviral drugs, no sufficient drug-level is reached in the patient, or the chosen drug-combination was not able to suppress the virus sufficiently. The latter occurs because the relations between observed mutations, phenotypic resistance and therapy success are poorly understood.
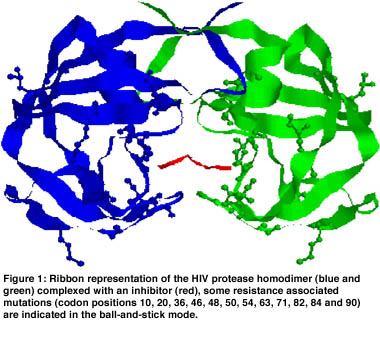
While the PR inhibitors, for example, all bind in the catalytic center of this enzyme, mutations associated with resistance occur at many different locations spread all over the three-dimensional structure of the protein (see Figure 1). The relation between point mutations and drug resistance remains unclear in many cases, not to speak of the interpretation of more complex mutation patterns.

The goal of the Arevir project is to develop bioinformatics methods that help to understand these connections and that contribute directly to therapy optimization. In a first step a database is set up in collaboration with project partners from university hospitals and virological institutes, in which clinical data, sequence data and phenotypic resistance data are collected.
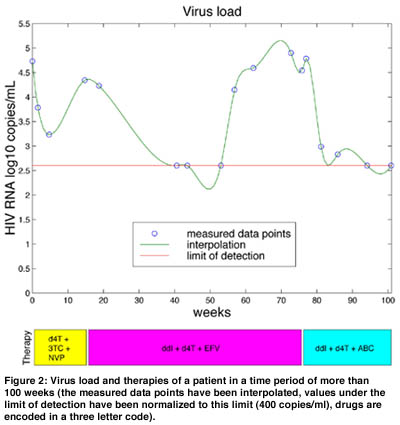
These correlated data are used to learn about the outcome of a therapy as a function of the drugs making up the components of this therapy and the genotype of the two relevant enzymes PR and RT. A successful outcome of a therapy can be measured as a substantial reduction in virus load (ie the number of virus particles measured in the patients’ blood plasma; see Figure 2). We can formulate a classification problem on the set of all pairs consisting of the therapy’s drug components and the amino acid sequence of PR and RT assigning to each such pair either the class ‘successful’ or ‘unsuccessful’.
We use decision tree building, a well known machine learning technique, to derive a model that can make an educated guess on the therapy outcome. The treating physician faced with the problem of finding a new therapy can consult the decision tree with a therapy proposal, provided a genotypic test result is available. In this way the automated interpretation of sequence data can directly improve patient care. Furthermore, the learned rules derived from the decision tree can be interpreted by virologists and clinicians, and help to better understand and manage drug resistance.
In future work the mechanisms of drug resistance will be studied at the molecular level. To this end we will carry out force field based calculations on enzyme-inhibitor complexes.
While traditionally bioinformatics methods are used in the course of developing new drugs, we develop and apply methods that can directly improve patient care by contributing to therapy optimization. Thus, in treatment of HIV infected patients techniques of interpreting genotypic data are of immediate relevance to therapy decision.
Links:
Bioinformatics at http://www.gmd.de/SCAI
Please contact:
Niko Beerenwinkel - GMD
Tel.: +49 2241 14 2786
E-mail: niko.beerenwinkel@gmd.de