Computerized Anatomical Segmentation of the Liver from Helical CT Scanners
by Luc Soler, Gregoire Malandain, Hervé Delingette and Johan Montagnat
Developed in the EPIDAURE group of INRIA Sophia Antipolis in collaboration with Prof Marescaux's team at IRCAD, Strasbourg, this work is a part of the European Eureka Project MASTER intended to create tools for planification and simulation of liver surgery. We present a new method to segment the eight liver's segments from portal vein with respect to Couinaud's anatomical segmentation, from helical CT scanners. These segments are used for the planification of segment-oriented liver surgery to localize the lesions and to define the cutting planes.
The liver's anatomical segmentation defined by Couinaud in 1957, describes its vascularization in eight segments. Each represents an area of influence of portal vein branches. The physicians say that when a tumor is localized in one segment, the whole segment has to be removed. The liver's anatomical segmentation is therefore a crucial information in liver surgery that allows precise localization of tumors and definition of cutting planes (see fig. 1).
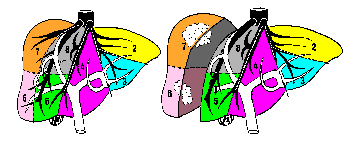
Figure 1: Definition of liver's segments (left) and sample of liver
surgery planification (right).
In order to build liver's anatomical segments, we first need to extract the portal vein. So we use helical CT scans with a 2mm thickness performed 50s after injecting a contrast agent by intravenous (portal time). We then reduce the image to the region of interest with the liver's mask computed by a deformable model techniques using local and global constraints. The resulting image can thus be divide in three classes, the liver tissue, the vessels with a higher grey-level and the eventual lesions appearing darker. Using this remark, we perform a three steps algorithm that automatically segment the portal vein from abdominal CT scans. The first step realize an automatic thresholding by computing threshold of vessels but also of potential lesions. The second step improves this result by a local analysis using a distance map. Finally, the third step removes false positives due to the anisotropy of images by performing a topological and geometrical analysis of the skeleton computed from the resulting vascular tree. This last analysis allows us to segment automatically the portal vein with all topological information as bifurcation points useful for labelling each vessels. Results on 10 patients showed that the algorithm automatically extracts the first three principal bifurcations of the portal vein, as well as can be achieved by manual segmentations. They also demonstrated that the skeleton analysis corrects errors of the first segmentation and incorrect vessel connections introduced by 3D reconstruction of the anisotropic images. This automatic segmentation allows us to label each branches of the resulting portal vascular network. Encouraging results show that area of influence of each branches give us a good first approximation of liver's segments (see fig. 2).
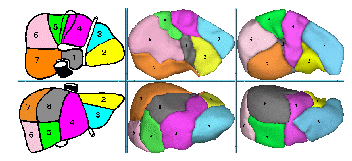
Figure 2: Comparison between atlas model (left) and two results computed
with our method.
We will now try to improve images by adapt the portal time to each patient. A good analysis will allow us to have better images with the same modality, and to obtain better segmentation. We also work on automatic labelling each vessels in order to automatically compute the anatomical segmentation. Another purpose is the automatic segmentation of tumors. First results using a similar method give here goods results, but it must be adapt to this specific problem. At least, cutting plane could be then computed, offered thus a complete tools for planification in segment-oriented liver surgery.
Please contact:
Luc Soler - INRIA Sophia Antipolis
Tel: +33 4 93 65 79 95
E-mail: Luc.Soler@sophia.inria.fr
