Magnetic Resonance Data Processing
by Piero Barone and Giovanni Sebastiani
Magnetic Resonance (MR) in biology and medicine is the main application field of the Signal and Image Processing group of the 'Istituto per le Applicazioni del Calcolo' (IAC) in Rome, the largest applied mathematics institute of the Italian National Research Council (CNR). In the last ten years, a number of collaborations between the IAC group and outstanding MR laboratories have focused on the development of models and methods for MR data processing. The activities include enhancing the resolution of MR spectra used for tumour identification, the quantification of MR signals from selected metabolites in brain pathologies, as well as MR image quality improvement, MR image reconstruction, dynamic MR imaging for human brain and lung pathologies, and functional MR imaging for the characterization of the areas of brain functions. Models and methods have been developed within either a classical or a Bayesian statistical framework, depending on the specific problem.
The Magnetic Resonance technique enables us to acquire digital data on the chemical and physical structure of a large variety of systems, including living systems. MR is increasingly used for both routine investigations and research purposes due to its very low degree of invasiveness. The analysis of MR spectroscopy data makes it possible to identify and quantify the chemical compounds in a given sample. MR image data processing provides tomographic slices as well as three-dimensional pictures describing the interior of physical objects.
Standard methods are available for MR data processing. However, such methods generally present serious drawbacks, eg they often have limited power as they do not always take into account information on both the physics of the MR data acquisition and the quantities to be estimated. Moreover, they are based on local optimization procedures. The aim of our current research is thus to provide more powerful methods that overcome these limitations.
From the physics of the MR data acquisition it is usually possible to derive a statistical model (the likelihood model) for the MR data, given the quantities to be estimated. However, the estimation of the quantities of interest very often involves the solution of an ill-posed problem, eg the solution of a Fredholm integral equation, the estimation of parameters of non-linear models.
In order to solve these problems, we have either developed robust methods based on highly sophisticated mathematical procedures, and/or we have incorporated into a Bayesian model some additional prior information about the quantities to be estimated. The a priori models we developed within the Bayesian framework, belong to the Markov random field model class. This choice has the advantage of reducing the computational complexity of the algorithms involved in the estimation. Special attention has been devoted to the problem of selecting the so-called hyper-parameters of the Bayesian model. In fact only by solving this problem is it possible to exploit the potential of the Bayesian approach for image and signal processing.
The methods we have developed include an original approach for solving super-resolution problems that permits the identification of unknown chemical compounds even when the noise affecting the data is moderately large. This method is based on simple statistical principles coupled with sophisticated complex analysis techniques. An example of application of this method is the identification and quantification of the beta-ATP region of in vivo 31P MR spectrum of human breast tumour implanted in mice, as shown in Figure 1. The dashed curve represents the Fourier spectrum, where the expected triplet is hardly recognizable. The solid curves are the lorentian lines of the triplet, obtained by estimating the spectral parameters from the MR signal.
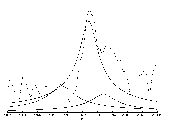 Figure 1: Beta-ATP region of
'in vivo' 31P MR spectrum of human breast tumour implanted in mice.
Figure 1: Beta-ATP region of
'in vivo' 31P MR spectrum of human breast tumour implanted in mice.
Another method estimates blood perfusion parameters from a temporal series of dynamic MR images acquired following the injection of a contrast agent. The method is based on local non-linear spatial filtering and on non-parametric linear regression. The method is simple and fast. Furthermore, it provides sufficiently accurate estimates of a few quantities, which are easily interpreted by the medical practitioner. An example of results using this method is shown in Figure 2. On the left of this figure, an MR image from a dynamic series of 30 images of the head of an ischaemic rat brain is shown. The oval region at the centre of the image is the brain and regional ischaemia appears in its left part (in white). The image on the right shows the result obtained by applying our method: the processed map of the blood perfusion defect in the interior of brain is superimposed upon the image on the left. The ischaemic region is better depicted in the processed image, where the two sub-regions called core (in white) and penumbra (in grey) also appear.
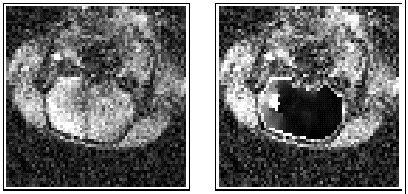
Figure 2: Dynamic MR imaging for diagnosis of ischaemia in the brain
of a rat.
This research has benefited from cooperations between the IAC group and the Physics Laboratory of the Italian National Institute of Health (Rome), the Department of Chemical Physics of the Weizmann Institute of Science, Rehovot (Israel), the Istituto di Strutturistica Chimica of CNR (Rome), the group of Statistics of the University of Tromsoe (Norway), and the MR-Centre in Trondheim (Norway).
Please contact:
Giovanni Sebastiani - IAC-CNR
Tel: +39 6 88470236
E-mail: sebast@iac.rm.cnr.it
